PLOS Computational Biology: GEMINI: Integrative Exploration of Genetic Variation and Genome Annotations
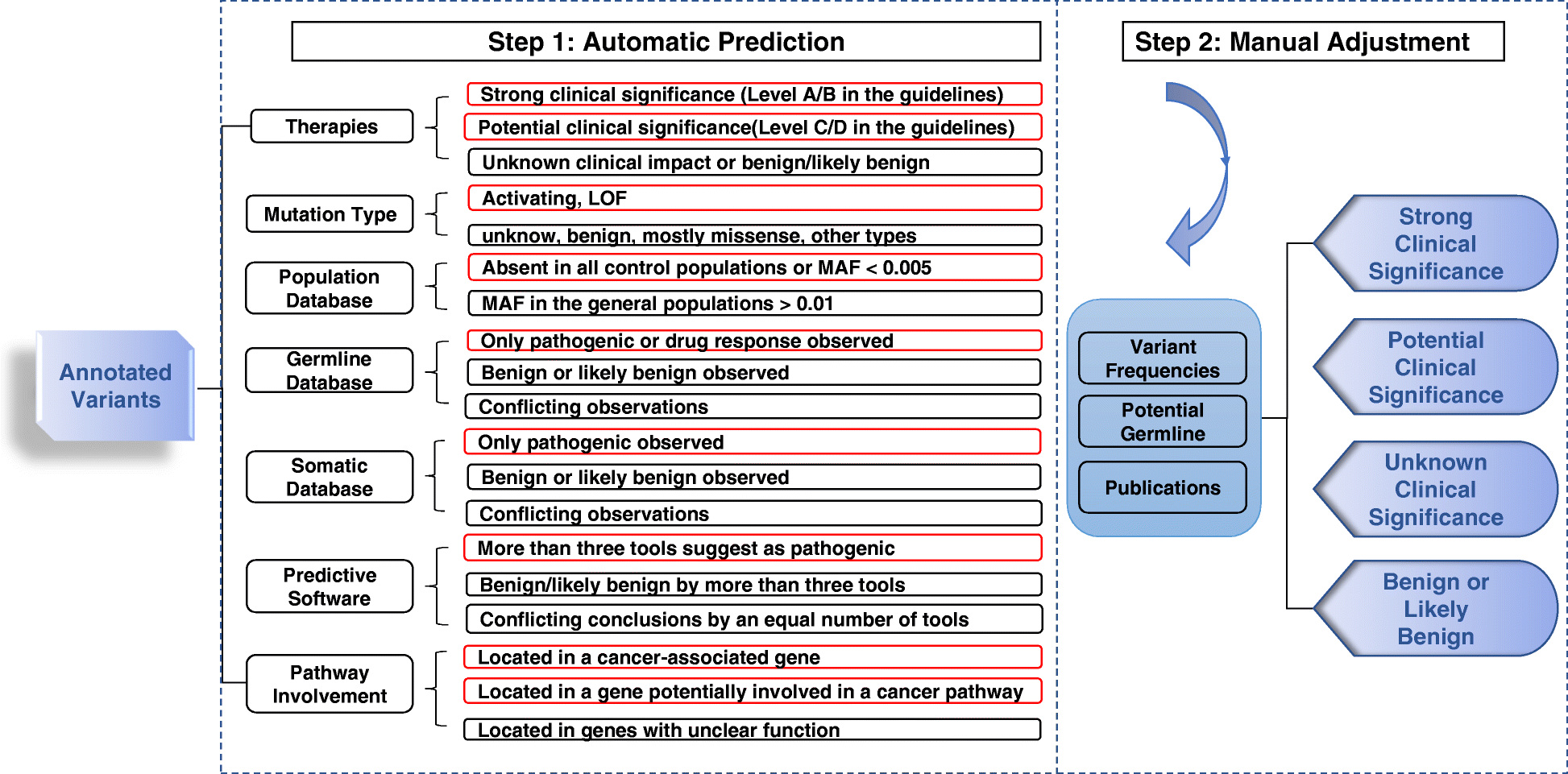
Variant Interpretation for Cancer (VIC): a computational tool for assessing clinical impacts of somatic variants | Genome Medicine | Full Text
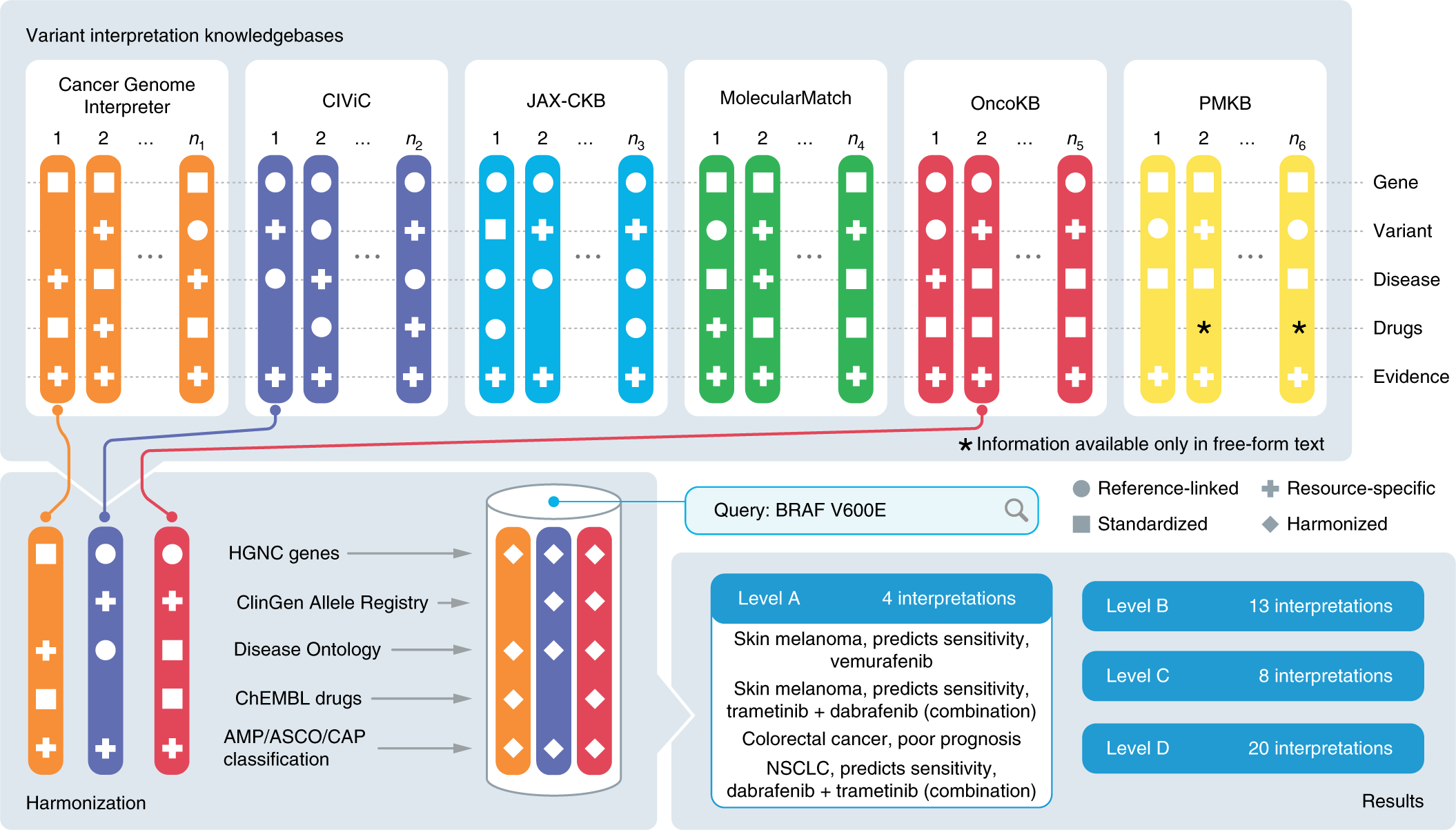
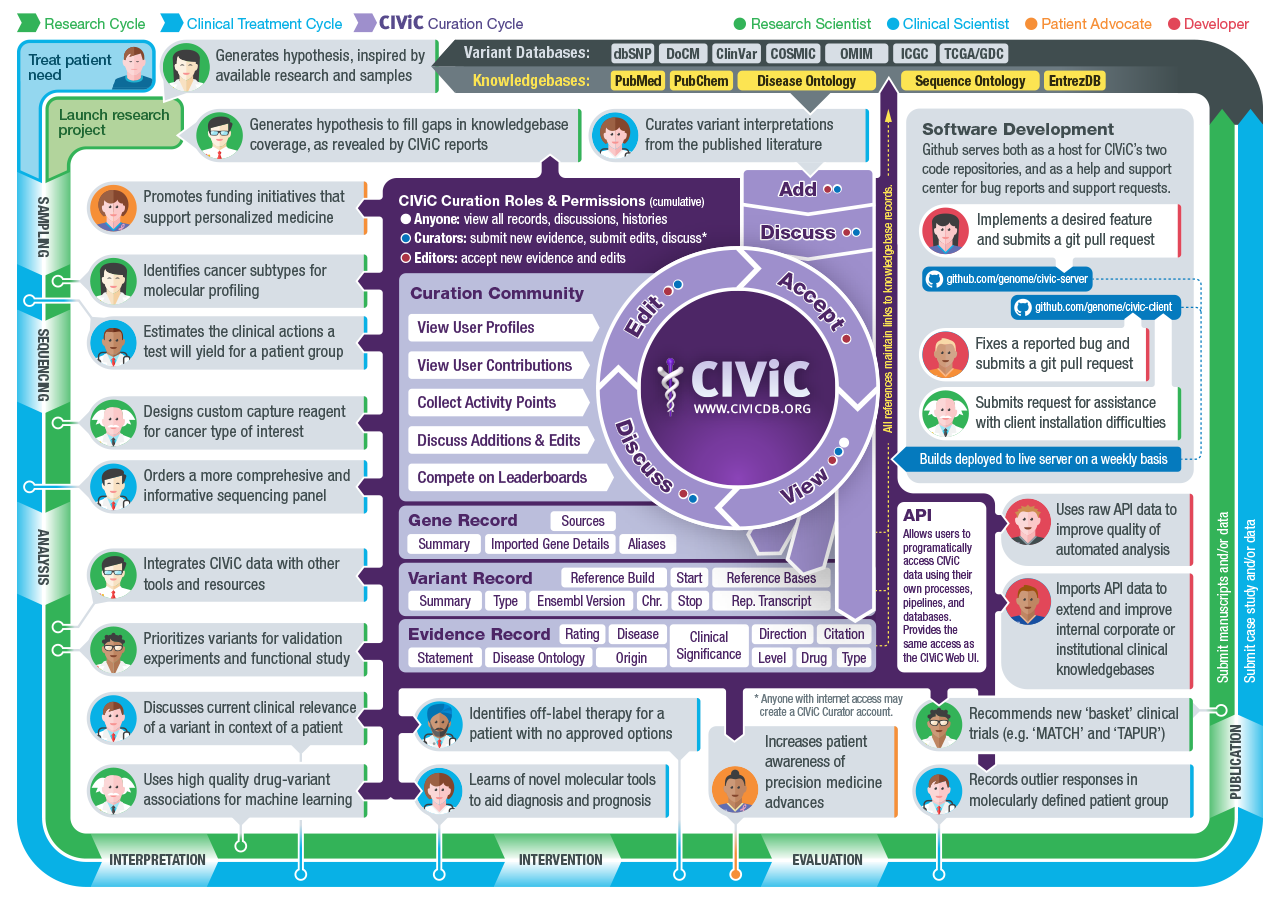
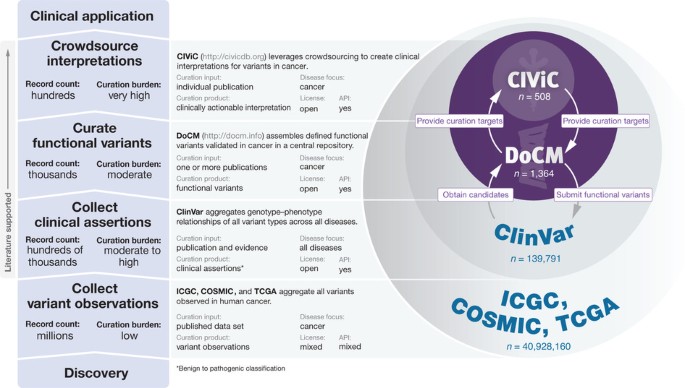
A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer | Nature Genetics

Technical desiderata for the integration of genomic data with clinical decision support - ScienceDirect
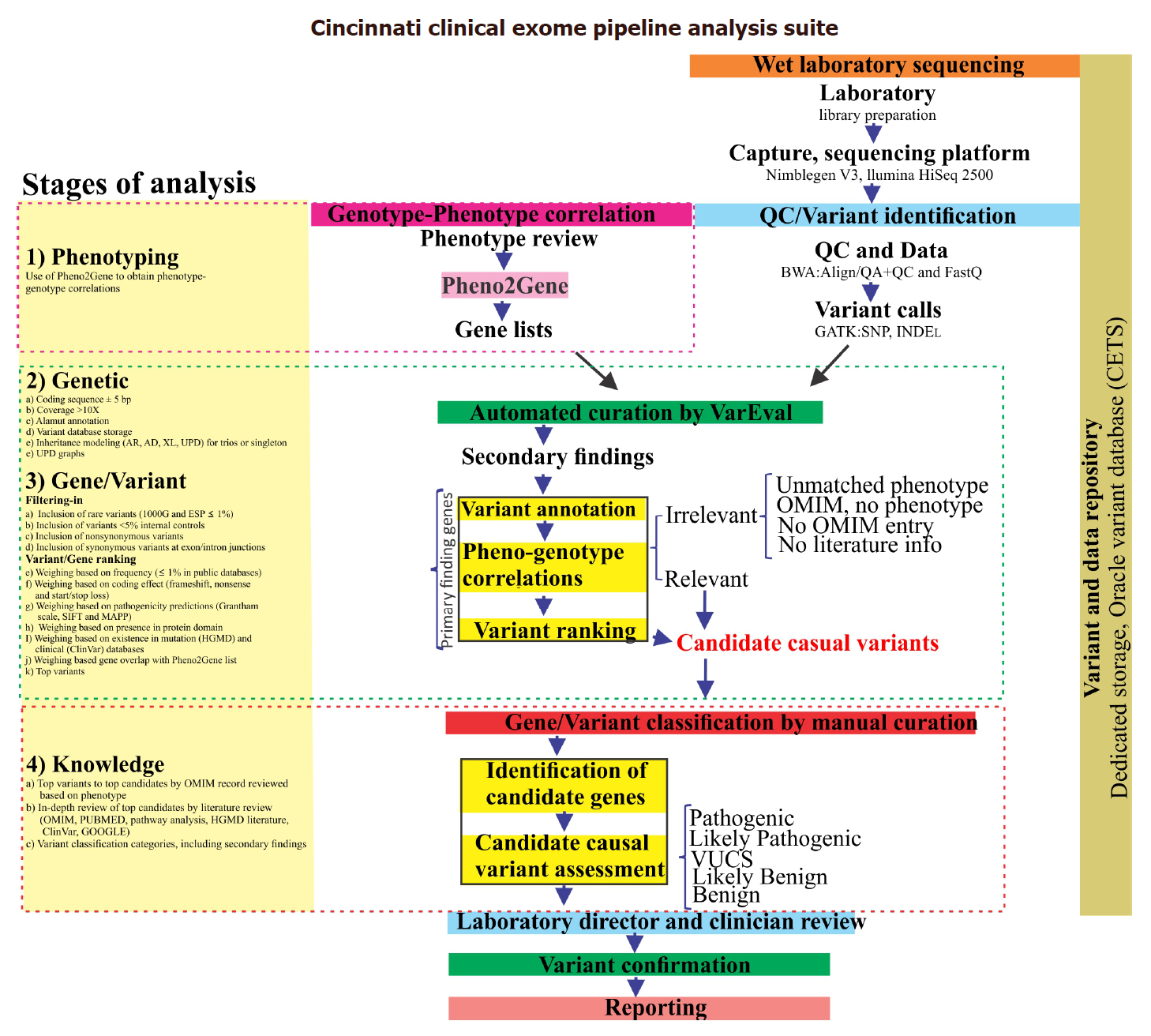
CCEPAS: the creation and validation of a fast and sensitive clinical whole exome analysis pipeline based on gene and variant ranking
HuVarBase: A human variant database with comprehensive information at gene and protein levels | PLOS ONE
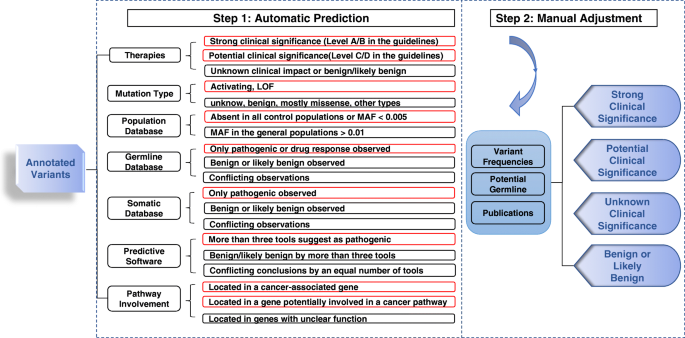
Variant Interpretation for Cancer (VIC): a computational tool for assessing clinical impacts of somatic variants | Genome Medicine | Full Text

Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer - The Journal of Molecular Diagnostics
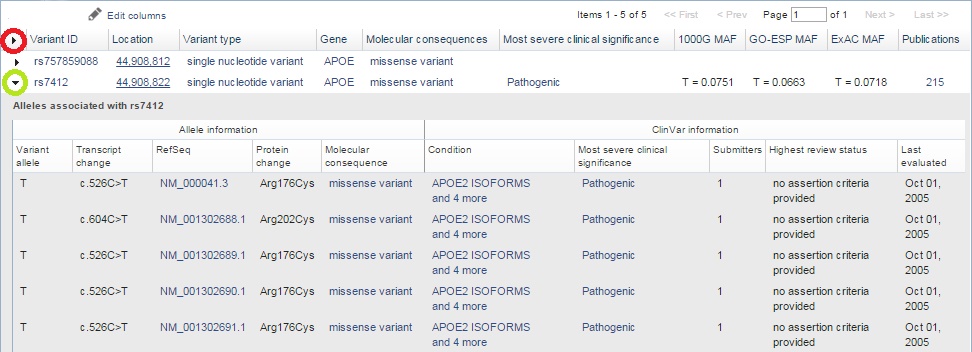
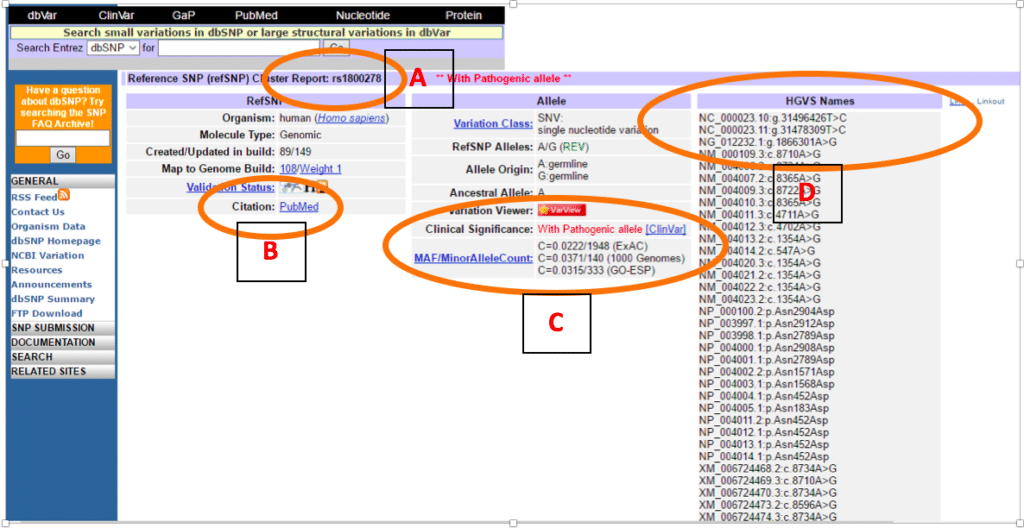
![PDF] Simple ClinVar: an interactive web server to explore and retrieve gene and disease variants aggregated in ClinVar database | Semantic Scholar PDF] Simple ClinVar: an interactive web server to explore and retrieve gene and disease variants aggregated in ClinVar database | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/253417a4bf8f7b320ab286223e372e99283d6dd4/3-Figure1-1.png)
PDF] Simple ClinVar: an interactive web server to explore and retrieve gene and disease variants aggregated in ClinVar database | Semantic Scholar

Standardized decision support in next generation sequencing reports of somatic cancer variants - Dienstmann - 2014 - Molecular Oncology - Wiley Online Library
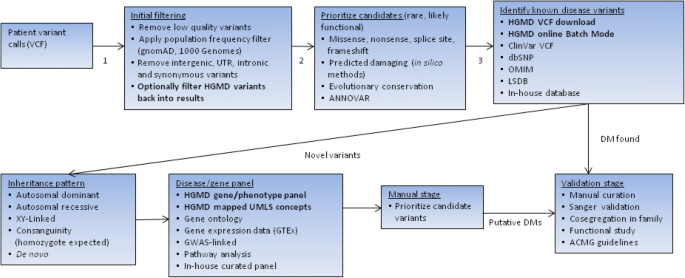
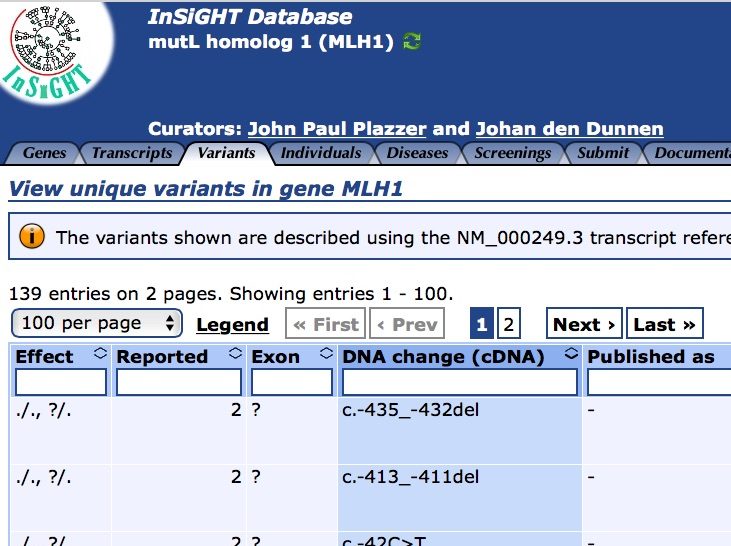
The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting | SpringerLink
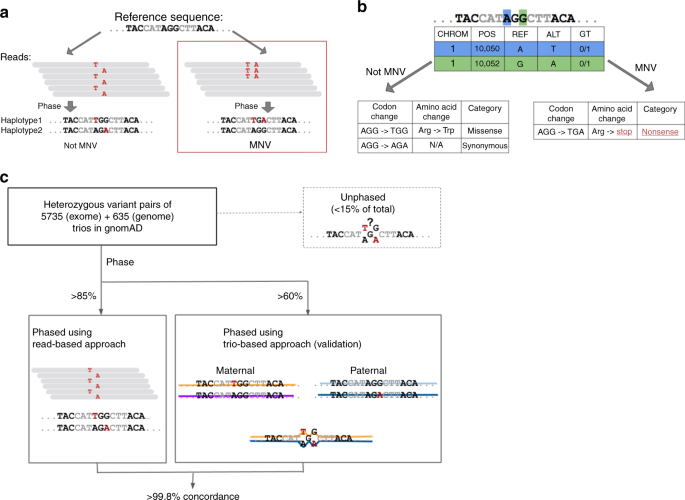
Landscape of multi-nucleotide variants in 125,748 human exomes and 15,708 genomes | Nature Communications